Stimuli-responsive hydrogel materials for wound healing
Infections associated with methicillin-resistant Staphylococcus aureus (MRSA), E. coli, P. aeruginosa, and Candida species have become a common problem in hospitals. Antimicrobial gels emerged as one of the strategies to prevent or treat such infections and have been designed in form of wound fillers, implant coating and for drug delivery. We designed a short 9-residue peptide that forms hydrogel in the presence of Ag(I) ions. Silver ions not only drive self-assembly of the peptide, but also provide antimicrobial properties to the material. The resulting gel demonstrates high potency against E. coli and S. aureus. Currently we design an antimicrobial hydrogel with the ability to change its stiffness in response to changes in redox state and pH.
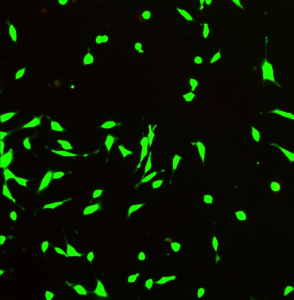
Below is an image of fibroblast cells (skin cells) in live/dead assay demonstrating that hydrogel is not toxic and could be used for wound healing applications.
Another useful property of our hydrogel materials is self-healing – hydrogel recovers its gelation state after removal of the strain. This property should allow for delivery of the preset hydrogel into any wound geometry using a syringe.
Inhibition of biofilm formation and biofilm dispersion of Streptococcus pneumoniae
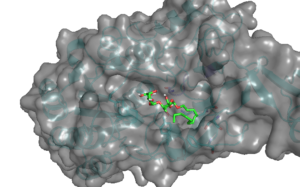
S. pneumoniae is a major human pathogen that causes pneumonia, meningitis, and otitis media that claim millions of lives every year worldwide being the top killer of children under the age of five. We identified a molecule that disperses S. pneumoniae biofilm at a relatively low concentration. We envision that this compound could be used in combination with antibiotics in a spray formulation that would eradicate biofilm, preventing tissue colonization and thus help treat pneumonia. Below is an image showing docking of identified molecule into its potential target on the surface of S. pneumoniae. We are working to identify the target of this molecule and to improve their binding.
NMR guided approach to evolution of Myoglobin protein
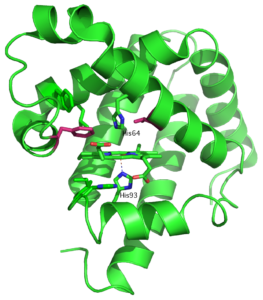
Proteins can be evolved to accept unnatural substrates or to catalyze new chemical reactions. The process of evolution is guided by available structural information or by computational analysis. We developed a new simple method for evolution of enzymes that relies on differences in HSQC spectra of the protein in the presence and absence of substrate analog. This method has been successfully used to improve catalytic efficiency of calmodulin-based Kemp eliminase that operates through acid-base mechanism. Here we demonstrate that our approach can be applied to other protein scaffolds and reaction mechanisms. Recent work by the Reetz group showed that Kemp elimination can be promoted by coordination of Kemp substrate to the Fe(II)-heme center of cytochrome P450-BM3 and subsequent electron transfer between iron and Kemp. To test applicability of NMR guided evolution to redox-catalyzed Kemp elimination we chose to work with heme-containing protein myoglobin. Based on NMR analysis we introduce mutations in the vicinity of heme and screen for accelerated formation of a yellow 2-cyano-4-nitrophenol product. Our work presents further validation of novel method developed for design of artificial enzymes.
Enzyme mechanisms
Ribonucleotide reductase (RNR) is an essential enzyme found in all organisms. The function of RNR is to catalyze conversion of nucleotides to deoxynucleotides. All RNRs rely on a metallo-cofactor to oxidize a conserved cysteine in the active site of the enzyme into a thiyl radical, which then initiates reduction of nucleotides. The proteins required for MnIII2-Y• cluster formation in class Ib RNR are NrdF (b subunit) and NrdI (flavodoxin). Oxidant is channeled from the FMN cofactor in NrdI to the dimanganese center in NrdF, where it oxidizes Mn2 center and the tyrosyl radical is formed. Both S. sanguinis and E. coli MnII2-NrdF structures have a constriction in the channel immediately above Mn2. In the E. coli, the constriction is formed by the side chain of Ser159, whereas in the S. sanguinis system it involves Thr158. This serine to threonine substitution is conserved in all streptococci, including human pathogens S. pneumoniae and S. sanguinis. Using stopped-flow kinetics we investigate the role of serine to threonine substitution in the mechanism of MnIII2-Y• cluster formation.
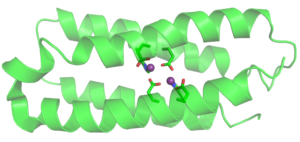
Design of mononuclear non-heme enzyme for oxygen activation
We incorporated a 2-His-1-carboxylate motif, often found in non-heme enzymes, into a four-helix bundle protein framework. Such coordination environment provides ligands to bind metal ions and leaves room to accommodate substrate, oxidant and cofactor. The resulting protein named FeLix is folded, thermally stable and binds a single Fe(II) ion. FeLix protein loaded with Fe(II) catalyzes oxidation of catechol by dioxygen, thus mimicking function of catechol dioxygeases, natural enzymes that have 2-His-1-carboxylate motif. One of the intermediates in the catechol dioxygenase mechanistic pathway is Fe(II)-semiquinone radical (SQ·) adduct. This intermediate has been observed in the Fe(II)- FeLix model system and characterized by UV-vis spectroscopy and EPR. We recently discovered that even apo protein can stabilize SQ• radical for weeks. We are working to understand what features of the protein are important for radical stabilization.